The first turn of our engineering crank 🛠🧂
Beyond deletions: additive strain engineering and adaptation
Introduction
Strain engineering, or optimizing a bacterium for a specific industrial process, is the bread and butter of biotech today. However, the majority of strain engineering focuses on deleting genes, rather than what bacteria in nature do—swap DNA to acquire useful traits horizontally.
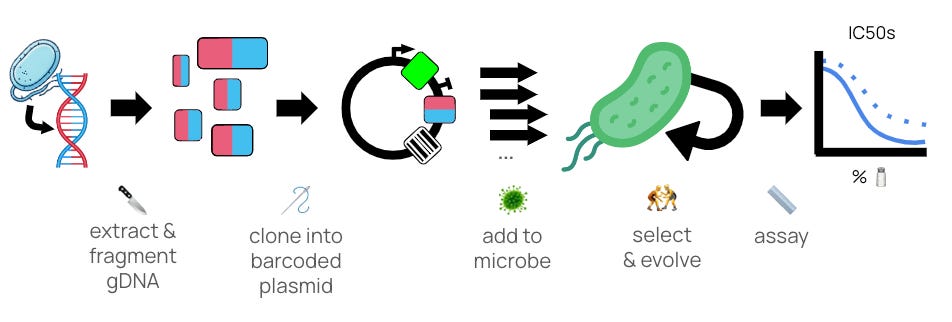
We built our ‘engineering crank’ 🛠 to mimic natural evolution by using functional genomics methods (like Boba-seq) to acquire new genetic material. DNA from an extreme organism is chopped up 🔪, reassembled into a plasmid library🪡, transferred into a recipient microbe 🦠, and selected with directed evolution methods (like the Long Term Evolution Experiment) for fine-tuning 🤼. At the end, we’ll measure whether or not the genes that stick actually improve growth in an extreme condition with a secondary measurement 📏.
To begin, we built and selected a first library of genes from salt-tolerant organisms in E. coli, and we’ll share those results here 🛠🧂. We hope this approach will allow us to engineer strains that are simply not possible with deletion-based techniques alone, including microbes for Mars!
🔪🪡 Making functional genomics libraries
An essential feature of our engineering approach is to mimic horizontal gene transfer by extracting DNA from a source microbe, putting it into a recipient microbe, and checking whether or not it changes the properties of the recipient. This sort of technique is called ‘functional genomics’ — not just looking at the sequence of DNA (genomics), but also what phenotypes it confers (function). Our goal here was to do a first proof of concept that uses functional genomics workflows to transfer properties related to Mars.
🧂Salt tolerance is one of the five extremophile properties a Mars microbe would need, and is perhaps the most well studied and easy to work with. We chose H. elongata as our DNA source of choice. H. elongata is a very salt-loving microbe: it was reported to grow in a range of high salt conditions up to 32% wt/vol, and we were able to reproduce its growth in salty media up to 16% wt/vol. It is also easy to grow and standard kits can be used to purify its gDNA.

Once we had extracted gDNA from H. elongata, the next challenge was putting that DNA into a recipient microbe 🦠. During this first proof of concept, we chose to insert DNA into E. coli, perhaps the most well-studied and easy-to-work with chassis. Techniques for inserting small pieces of DNA - not whole genomes - are more numerous and efficient, so the next step is to fragment the gDNA 🔪 and reassemble those fragments into a plasmid library 🪡.
🔪 Of the many ways to cut DNA, which one should we use? One common approach is to fragment gDNA by physical shearing with ultrasonication then clone it into a plasmid with blunt end ligation. This is great for generating libraries with ~104 to 105 members, but we would like to push toward libraries with 107 members for covering metagenomic samples. We found a paper that co-opted Tn5 transposition and tagmentation, more commonly used for preparing shotgun sequencing fragments, to make functional metagenomic libraries by Mosaic Ends Tagmentation (METa) assembly. They show higher transformation and cloning efficiencies of METa compared to blunt end ligation methods, and suggest that METa is compatible with lower amounts of input gDNA, which would be useful for low yield environmental samples.
We used the METa protocol to build a library of H. elongata gDNA using a commercially available tagmentase following the manufacturer's protocol. We tried the tagmentation reaction at several ratios of tagmentase to gDNA and used an Agilent Tapestation to check the products. This is followed by a gap filling reaction, assembly reaction, and transformation, with various DNA clean-up steps along the way. The end result is a protocol for robustly cutting gDNA into fragments of our desired size range 🔪.

Once we had fragmented gDNA, we needed to clone those fragments into a plasmid library🪡. We had previously constructed a barcoded empty plasmid with 107 variants, and used that as the backbone for our library. For our first time trying the METa protocol we used AarI Golden Gate Assembly and were able to generate two 106 barcoded H. elongata gDNA fragment plasmid libraries in NEB DH10β electrocompetent cells 🦠.
We submitted our assembled library for Whole Plasmid Sequencing by Plasmidsaurus, and the QC results are shown below. While we were happy with the library size, we were surprised to see that the inserts were relatively short given our input tagmented gDNA profile (mean >10kb), and thus very few of the inserts contained entire gene sequences.

How not to do it:
🔪Tagmentation and 🪡 Golden Gate Assembly were not the only techniques we tried! We actually tried four different assembly methods.
🫠 Golden Gate Assembly (AarI) | This was our first choice because of its high efficiency and low error rate. You may remember that to effectively use Golden Gate Assembly, you need to be sure the enzyme recognition site doesn’t exist in your sequence. That’s why we used AarI instead of BsaI - there was a BsaI site in our barcoded plasmid backbone. Unfortunately, after being confused by our sequencing results (shown above) for a while, we realized that when we digested our tagmented fragments to generate the AarI sites, we were likely also digesting the 1180 AarI sites (only 7bp long) present on the H. elongata genome and therefore removing larger fragments from our library 😭! Ironically, despite the fact that our first AarI-generated libraries are non-ideal, we still moved ahead with selecting it because it had enough inserts long enough to contain both promoters and fully intact genes.
Electronic lab notebook: First time: fragment genomes (public copy)
🤔 Gibson Assembly | Now we know why the published METa protocol used Gibson assembly for their libraries. We repeated our tagmentation protocol, but this time with Gibson homology sites appended onto the Tn5 ME sites as well as on our barcoded plasmid, and were able to get a 107 library. But when we got our sequencing data back, most of the transformants didn’t have an insert. Looking closer, somehow during the assembly process, we both 1) did not get fragment insertions and 2) part of the plasmid backbone near the insertion site got deleted in a way that let the backbone ligate to itself. While we could not find a clear reason for why this happened, it could potentially be fixed by shifting the Gibson assembly site and trying again. In the meantime, we decided to try two other options in parallel.
Electronic lab notebook: Add barcodes during PCR (public copy)
👎 Homing endonuclease (I-CeuI) | To avoid cutting up the gDNA, we looked into using a restriction enzyme with a larger recognition sequence. We decided on I-CeuI, which has a 27bp recognition site that does not exist in the H. elongata genome. We ordered new primers for transposome assembly with the I-CeuI recognition site in it, repeated the whole tagmentation protocol, and got a 105 library. Unfortunately, these also appeared to be mostly empty based on our sequencing results. Our hypothesis is that this older homing endonuclease might just have lower activity compared with other more optimized restriction enzymes.
Electronic lab notebook: Rare restriction enzyme (I-CeuI) for Tagmentation (public copy)
What ended up working:
🥳 Uracil-Specific Excision Reagent (USER) | In parallel, we also tried USER cloning, where we tagmented with primers that contained uracil. Then we gap-filled with a polymerase that can tolerate uracil and digested with a uracil-excision enzyme to make custom 10-base sticky ends. This yielded a library with 105 members and with larger insert sizes, and we may rely more on this technique moving forward.
Electronic lab notebook: USER for Tagmentation (public copy)
Future improvements:
While we are able to generate extremophile gDNA-sourced functional libraries, there are several areas in need of optimization that are ongoing and queued up for the future. We would appreciate any suggestions you may have!
🧬🧀 Extracting gDNA from other extremophiles and metagenomic (!!) samples
⚡🧫 Optimizing tagmentation → transformation protocol to generate larger libraries, compatible with metagenomes
🚀🧪 Mars-ifying our media beyond just salt
🔎🏔️ Bioprospecting for our next metagenomes
🦠🤼 Selecting libraries of horizontally transferred genes
Once we’ve prepared libraries of extreme DNA, we need to add them to our recipient microbe🦠, and challenge them to tolerate an extreme condition and see what survives 🤼. To that end, we took two replicate libraries we had made (with the AarI Golden Gate cloning method), and selected them in high salt to see if we could pull out any genes that enhanced salt tolerance in E. coli.
These libraries are not ideal. The maximum insert size is about 2.5 kB, and the mean is only about 1 kb. Also, the selection happened in the DH10β cells we built the libraries in instead of a more relevant strain. But our purpose here is to demonstrate that the crank works, not to build a super-salty organism in one try. We also only had two independent replicates of the library, so to ensure replicates we split each library in four, then selected for 7 days in salt.
It’s important to carry at least 100 million CFU through every single step, because otherwise we would ‘bottleneck’ our library and lose members because we didn’t take enough cells. We did some quick tests to ensure that the 500 uL of a glycerol stock and 100 uL of culture was sufficient for that, then went ahead with the experiment described in the figure below.

How not to do it:
Our first attempt failed because the salt concentration was too high. We chose LB + 5% salt because DH10β seemed to grow reasonably well in 5% salt on the first day. But alas, the day 2 cultures grew significantly worse, and the day 3 cultures didn’t grow at all. If you select a population to stress that is too great, it will go extinct, not evolve! 🦖 Lesson learned.
What ended up working:
We tried a few other stringencies with one of our library replicates and saw that LB + 4% salt retained growth over several days. So we restarted the experiment from the intermediate glycerol stocks and selected 4% salt, which went smoothly.
Future improvements:
In the future we plan to move to plate-based selection methods, potentially using 24 or 48-well plates to increase volumes. This would make experimental setup easier and would let us try multiple selection stringencies at the same time. This would both ameliorate the risk of killing everything with too harsh a selection and allow us to compare how the results differ among different selection pressures.
📏 Did it work?
Once we had our selected libraries, it was time to see if we’d managed to select for salt tolerance. First, we spot-checked some cultures from the last day of selection by miniprepping them and submitting to Plasmidsaurus for Whole Plasmid Sequencing. Then we checked the raw reads and saw that diversity was pretty low - with only one or two variants dominant in each culture.
With confirmation we’d selected for something, we turned to our handy IC50 assay to measure salt tolerance 📏🧂.
How not to do it:
First we ran the IC50 experiment directly on the cultures that emerged from the selection, comparing them against the plasmid expressing an mStayGold (a negative control that should not improve salt tolerance) and the Dr-IrrE (a positive control gene known to improve salt tolerance) in the same strain. While the data looks promising, we can’t conclude much. There could be SNPs in the genomes of these strains that are independently increasing salt tolerance, and we aren’t even certain if they’re clonal or still populations of multiple library members (hint, they’re populations). You can see the lab notebook for this experiment here: IC50 of 1st selection experiment winners (public copy).
What ended up working:
The right thing to do here is to re-transform our winners into a clean background so we can tell how much of the effect came from our plasmid. To do this we miniprepped all of the ‘winning’ cultures - which may still be mixed populations - and transformed them into the K-12 wild-type E. coli, then sequenced colonies. We found 7 unique genes, and tested a few replicates of each in our IC50 assay.

Not only do these genes successfully transfer salt tolerance from H. elongata to E. coli, when we look them up, biologically their mechanisms of action largely1 make sense!
Dr-IrrE - a regulatory metalloprotease. This is our positive control.
fpR/trapT - a ferredoxin-NADP(+) reductase. Indeed, fpR mutants are salt sensitive.
HELO_4410A - Ankyrin repeats. Perhaps important in plant drought resistance?
DUF1486 - a 3-beta hydroxysteroid dehydrogenase. Indeed, steroids are associated with microbial salt balance regulation.
hupB/lon - a histone-like DNA-stabilizer. These are important stress-response protein in mycobacteria.
BetG - a betaine/carnitine transporter. These are osmoprotectants
HELO_2306 - Serine protease? Not sure, mostly unannotated.
opuE - a Sodium/proline symporter which works well in high salt to acquire nutrients.
In summary, we were able to successfully isolate some fragments of the H. elongata genome that increased salt tolerance in this assay.
Future Improvements:
Next time we probably won’t bother with checking the salt tolerance of the selection cultures, and will go straight to cloning into a clean background. We’re also continually improving the throughput, ease and accuracy of the IC50 assay, and I’m hoping we’ll be able to test even more variants soon.
What’s next?
This first experiment has laid the groundwork for showing that we can simulate horizontal gene transfer between microbes and use it to successfully ‘transfer’ a Mars-relevant property from one microbe to another. Making a Mars microbe that can survive many extreme conditions at once will require applying this technique many times to grab extreme properties from many different organisms 🦠 🚀.
Here are a few next steps that we’re excited about.
In addition to selecting on salt we could select in high perchlorate. Can we make a salt-and-perchlorate tolerant microbe 😵 🧂? This would be valuable for biomanufacturing on human missions to Mars.
Which organisms should we source DNA from? We now have a wide variety of interesting libraries now with larger insert sizes and from more exotic extremophiles and even microbial community samples 🧫. We’re trying to decide which ones to select next!
Eventually we want to iterate this library selection, to build up tolerance by combining genetic material from many different sources into one organism. We’ve got a couple (broad host-range) methods for this under development! 🛠
All of the library members were barcoded 🏷️ and we’ve done a sequencing run on them! It’s about a billion paired-end reads. Analysis of fitness effects 🏋️ in progress! If successful, this gives us an opportunity to gather big data on the properties of millions of genes in a single assay.
💬 Do you have opinions?
We want to get feedback on our work faster than traditional scientific publishing will allow. That’s why we’re posting our science updates here. Please do comment below if you have thoughts, on this post or the attached lab notebooks, and sign up to get future updates!
HELO_2306 & opuE didn’t improve salt tolerance in our IC50 assay. Why not? We think our salt selection and the salt tolerance assay aren’t exactly overlapping in what they measure. These genes might help with serial culture in high salt in a way that’s invisible to our IC50 assay. Interestingly, opuE only came up very late in the selection - it appears to not be selected for early, but be able to outcompete some of the other salt-tolerance genes over the long run. Given past literature it seems like it might be more important for efficiently acquiring nutrients in high salt conditions, which suggests maybe its primary benefit is competition against salt-tolerant strains. HELO_2306 is a barely-annotated serine protease that could be doing almost anything, so it’s hard to say much.
Cite as: Pioneer Labs Reports (2025). The first turn of our engineering crank. figshare. Online resource. https://doi.org/10.6084/m9.figshare.29042423.v1







Cool work! Have you tried Gateway for library assembly? You could add attB sequences instead of Gibson homology sequences, although you'll need to modify your plasmid to be compatible. Gateway (or the MegaGate variant) has worked for me to clone libraries.
Of course if you're happy with USER then you can just keep using that.
Which organism to source genes from? I would suggest Cryptococcus neoformans fungus. It thrived in the Chernobyl wastelands, it loves gamma-radiation.
https://pubmed.ncbi.nlm.nih.gov/27899501/